install.packages("nycflights13")
install.packages("dplyr")
install.packages("knitr")
library(nycflights13)
library(dplyr)
library(knitr)Tutorial 2 - Navigating RStudio Tutorial: Using R Scripts and R Markdown Files
Create a GitHub Repository for Tutorial 2
Before we start with our coding exercises, we’ll want to create a new GitHub repository for Tutorial 2. Follow the instructions listed in Tutorial 1 to create a new GitHub repository called hpam7660-tutorial2.
Open a new R Project
Navigate to the File -> New Project tab in RStudio. Let’s call this project “Tutorial 2”. Follow the instructions from Tutorial 1 for linking the new project to your newly created GitHub repo.
RStudio Window
Let’s take a minute to briefly go over the RStudio interface. When you open RStudio, you’ll typically see the following three windows.
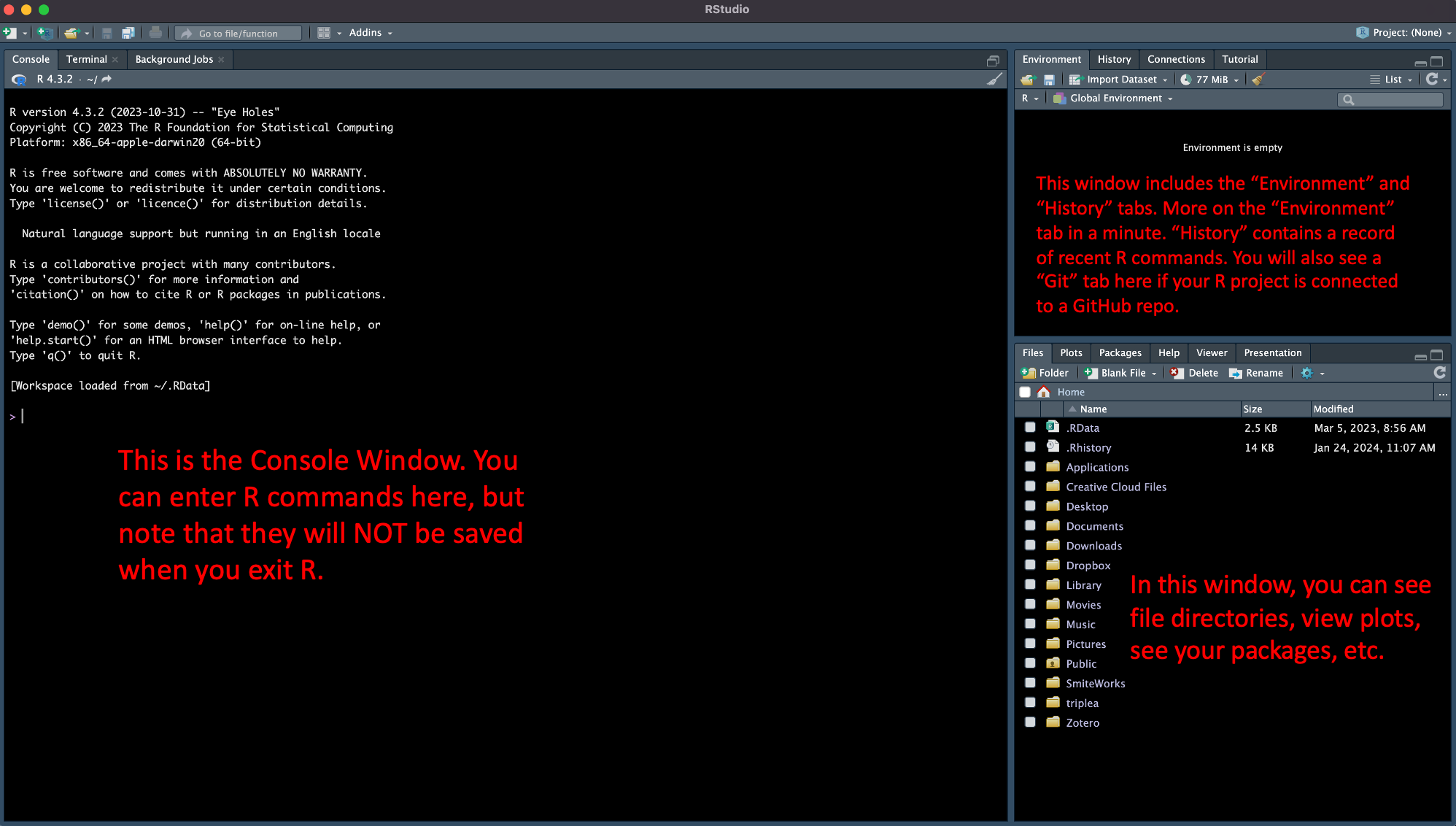
There’s a lot more here, but we’ll cover other aspects of RStudio as they come up in our tutorials and problem sets.
Console Command Line
Now that we have a new project linked to the proper GitHub repo, let’s start by running a few commands in the RStudio Console Window command line. This is the window at the bottom of RStudio (make sure the “Console” tab is selected). Throughout this tutorial, we’ll be working through the examples in Chapter 1.4 of ModernDive.
First we need to install and load the packages we’ll need for the tutorial (you may already have some of these packages installed, but it won’t hurt to reinstall them). Type each of the following commands in to the RStudio command line:
Exploring Data Frames
Now that we’ve loaded the packages and libraries, let’s take a look at the data objects contained in nycflights13. To do so, click on the dropdown icon next to “Global Environment” and select “package:nycflights13”.
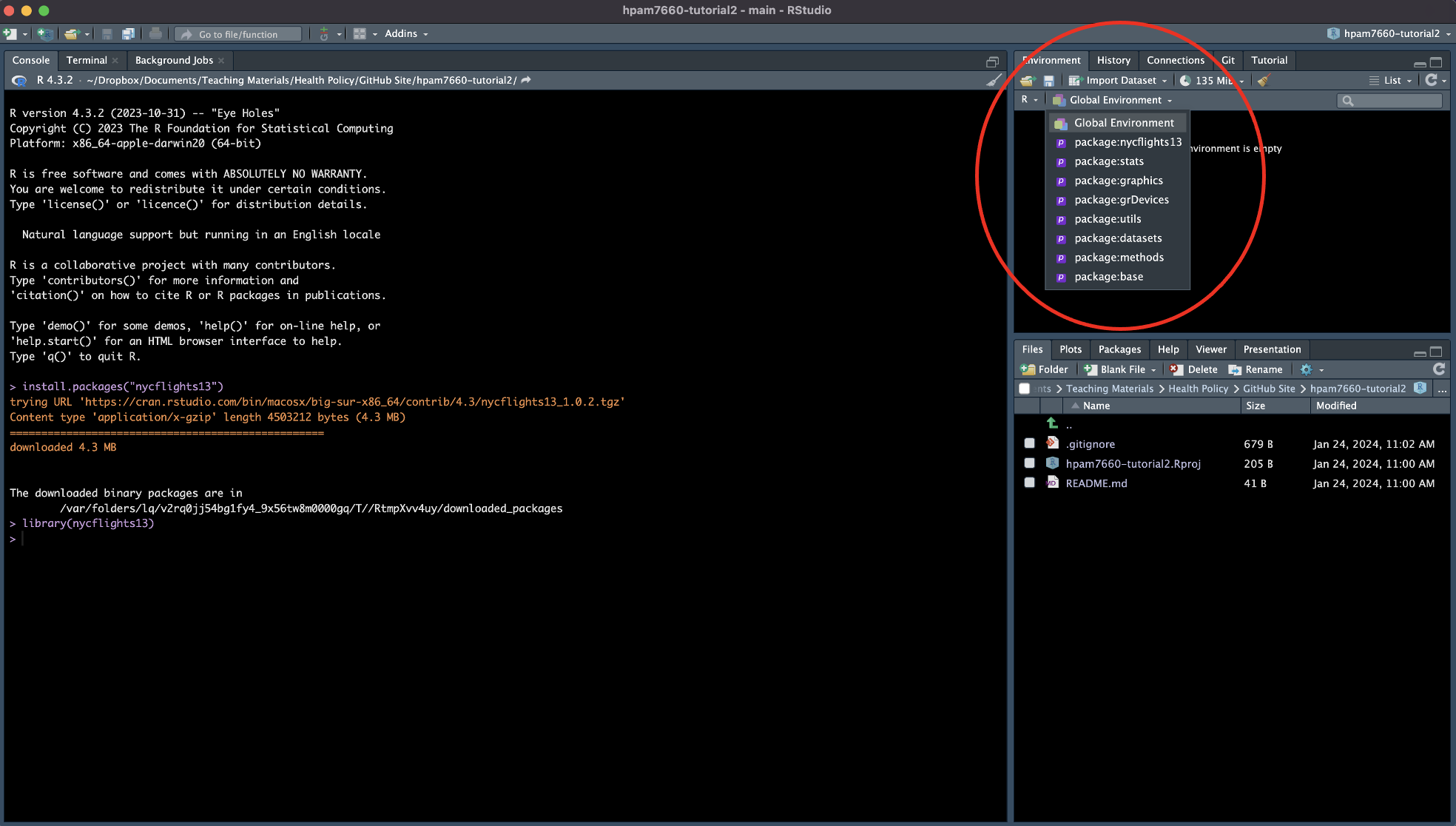
You should now see a series of objects in the “Environment” window.
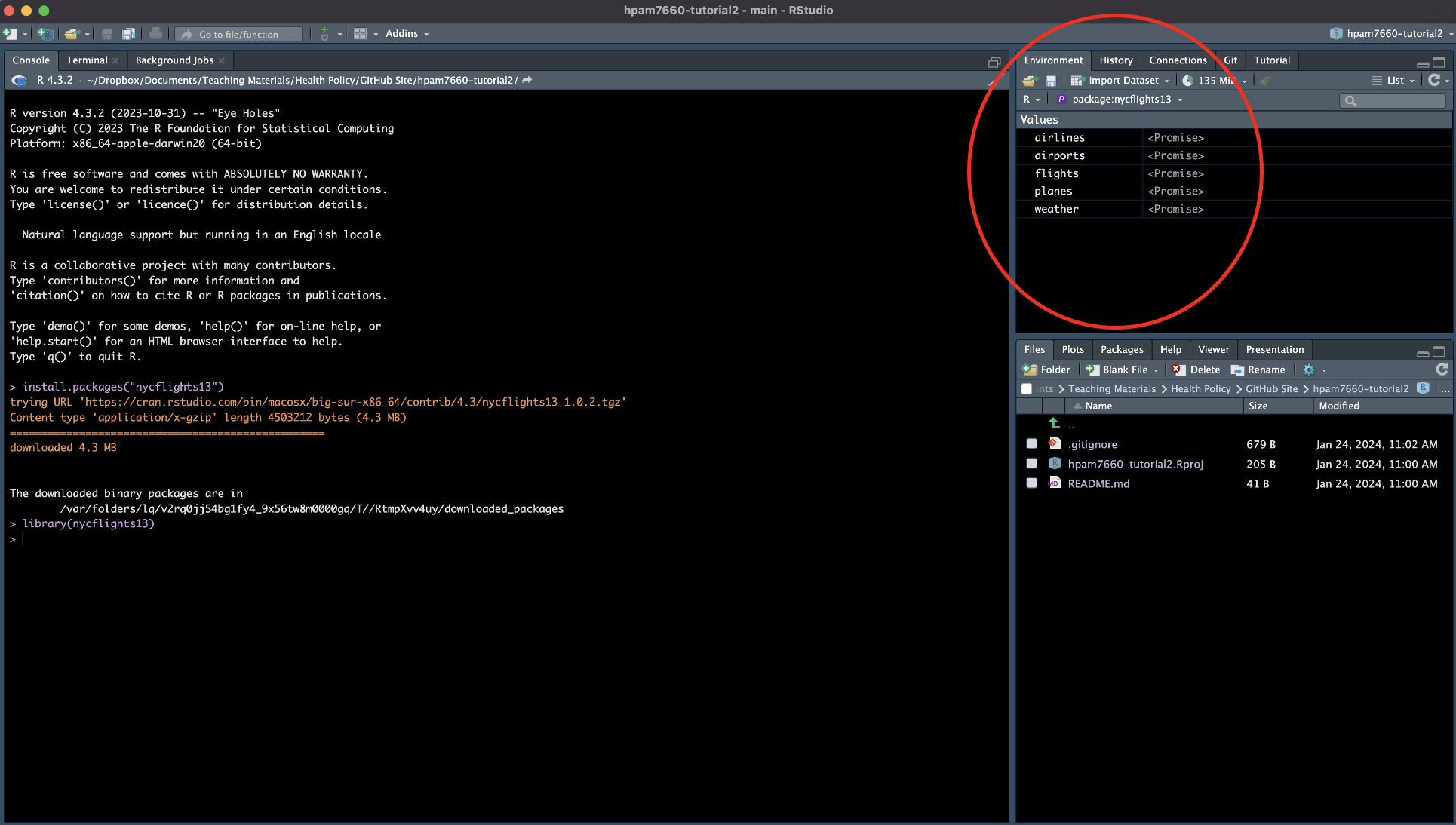
Let’s take a look at each of these objects to see what information they contain. Type the following code into the Console Window command line and hit enter:
flightsYou should now see a “tibble” that displays the first 8 columns and the first 10 rows of the flights data frame. The tibble also tells us that the data frame contains 336,766 more rows (beyond those displayed in the Console Window) and 11 more variables/columns (each of which is listed).
There are several other ways that we could examine the flights data frame including:
- Using the
View()function to open RStudio’s built-in data viewer. This allows us to scroll through all columns and rows in the data frame. - Using the
glimpsefunction, which is part of thedplyrpackage. This is similar to the tibble that we saw by simply typingflightsinto the console, but also includes each variables data type (e.g., integer, double, character, etc.) - Using the
kable()function, which is part of theknitrpackage. This function is helpful when we want to generate formatted output in an RMarkdown document, but it’s important to note thatkablewill print the entire data frame by default.
Let’s try some of these out. Use the glimpse function to take a look at the flights data frame and the kable function to examine the airlines data frame.
Data Manipulation
Now let’s make some manipulations to the data. Suppose we want a list of flight numbers and delay times for each United Airlines flight that was delayed by 4 hours or longer in January 2013. First, we can subset the data so that we only retain United Airlines flights. We can do this using the carrier variable since the code “UA” in this field identifies United Airlines flights. Go ahead and type the following into the Console command line and hit enter:
ua_flights <- filter(flights, carrier == "UA")Notice the new data object in the “Environment” tab called “ua_flights”. Let’s take a look at it using one of the viewing methods above (use whichever you prefer).
Now we need to determine which UA flights in January 2013 were delayed by 4 hours or longer. We can do another subset of the data using the year, month, and arr_delay variables (notice that the arr_delay variable is measured in minutes). Type the following into the Console command line and hit enter:
ua_delay <- filter(ua_flights, year == 2013, month == 1, arr_delay >= 240)Now we have a data frame that includes all UA flights that were delayed by 4 hours or longer in January 2013. Notice that we didn’t need a separate filter command for each argument (i.e., year, month, and arr_delay), we simply combined them all into a single filter command.
Finally, since we’re really only interested in the flight numbers and delay times, we can get rid of the extraneous variables in the data frame using the select command. Also,
ua_final <- select(ua_delay, flight, arr_delay)Now that we have our final dataframe, let’s take a look at the table of flight delays using the kable function:
kable(ua_final)Using R Scripts
So far so good, but what happens if we were to close out of RStudio? Or, say 6 months from now, we decide we want to re-generate the table we just made? Entering commands directly into the Console Window command line is not a very good way to structure our workflow because once those comments are gone, they’re gone for good! A better way to track our work and ensure that we can reproduce our results if necessary is to use an R script file to write our code. Let’s take a look.
Navigate to the File -> New File -> R Script tab in RStudio. This should open a new window in RStudio called the “Source” window and a document called “Untitled1” where you can write and save your R code.
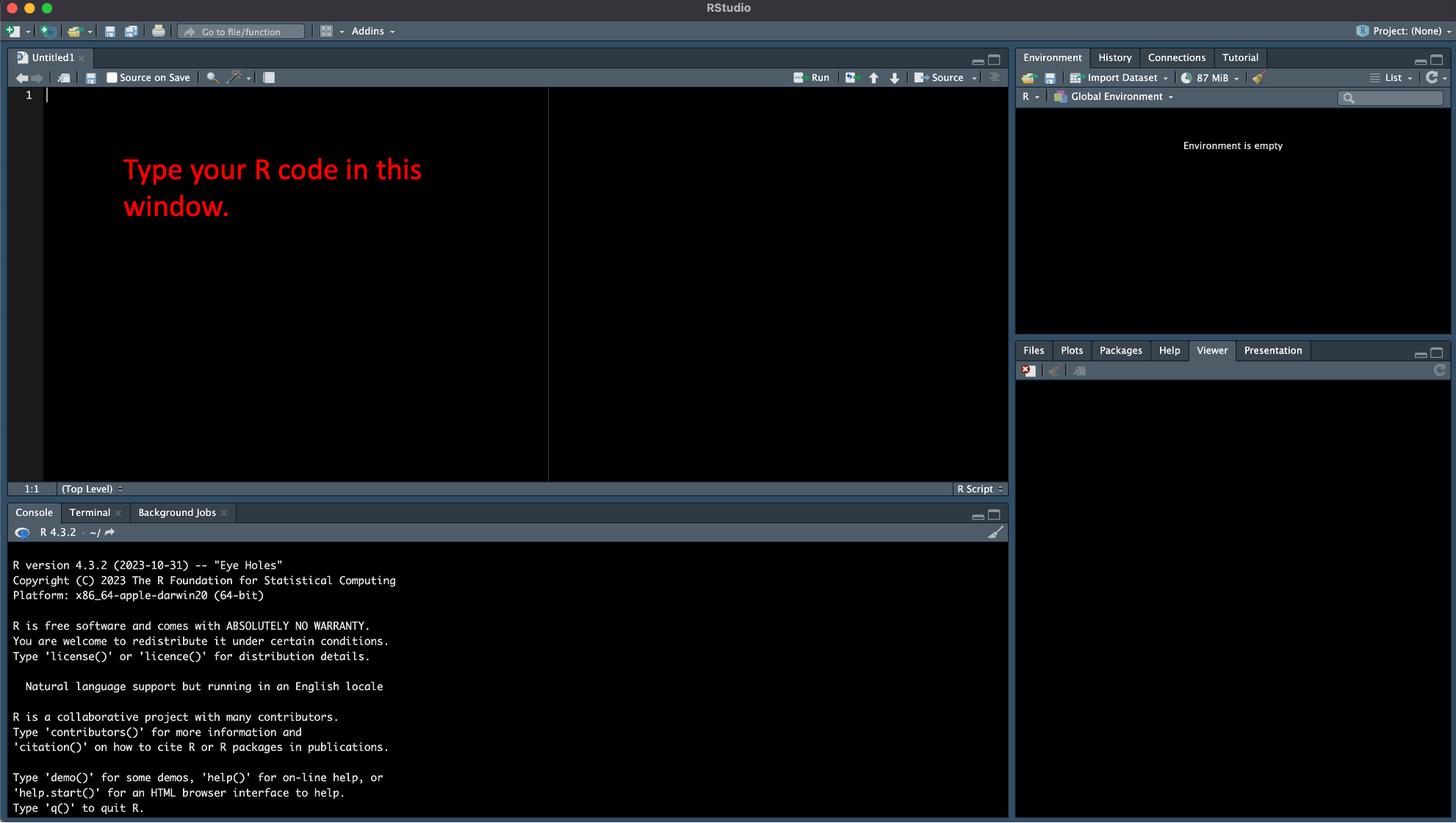
Let’s give this file a name and then recreate our United Airlines flight delay table by writing the code we entered directly into the command line into our R Script.
To name the file you can click on the save icon in the menu bar or navigate to File -> Save. Either way, you should be prompted to name the file (which will have a .R extension) and choose a location for saving. Let’s call this file Tutorial_2 and save it in the folder you created for your new hpam7660-tutorial2 GitHub repo. Notice that if you click on the “Git” tab in the upper righthand window, you should see this new R script ready to be staged, committed, and pushed to the GitHub repo.
Now type the code below into the R script (note that I have combined a couple of lines from the previous version):
library(nycflights13)
library(dplyr)
library(knitr)
ua_delay <- filter(flights, carrier == "UA", year == 2013, month == 1, arr_delay >= 240)
ua_final <- select(ua_delay, flight, arr_delay)
kable(ua_final)Once you’ve typed this into your R Script, save it, and click on the “Run” button in the menu bar. You should see the code run and the output table displayed in the Console window. Now if you exit RStudio, you’ll have the script file that you can run at a later date and reproduce the exact same table. Further, once you push the script file to GitHub, you can share it with other collaborators who may be working with you on the project.
We’ll push the R script to GitHub in a few minutes, but first let’s try using an R Markdown document to track our coding and it’s associated output.
Using R Markdown Documents
To open a new Markdown document navigate to File -> New File -> R Markdown. You should see a popup box with a button in the lower left corner that says “Create Empty Document”. Click that button. We could choose another formatting option, but RStudio will include a bunch of stuff that we don’t need in the final document. You should now see an RMD tab called “Untitled1” at the top of the Source Window. Click over to that tab.
Markdown documents always begin with something called a YAML header. The YAML header allows us to add things like a title, author name, date, etc. to the top of our output document. Add the following code (using your name) to the top of your Markdown document:
---
title: "Tutorial 2"
author: "Kevin Callison"
date: "February 1, 2024"
output: pdf_document
---Note that we also tell Markdown the type of output document we want in the YAML header. In this case, we’ll go with a PDF, but other options include an html_document or a word_document.
Now let’s add the same code we used in our R Script file:
library(nycflights13)
library(dplyr)
library(knitr)
ua_delay <- filter(flights, carrier == "UA", year == 2013, month == 1, arr_delay >= 240)
ua_final <- select(ua_delay, flight, arr_delay)
kable(ua_final)To generate the output PDF file, click on the “Knit” button.
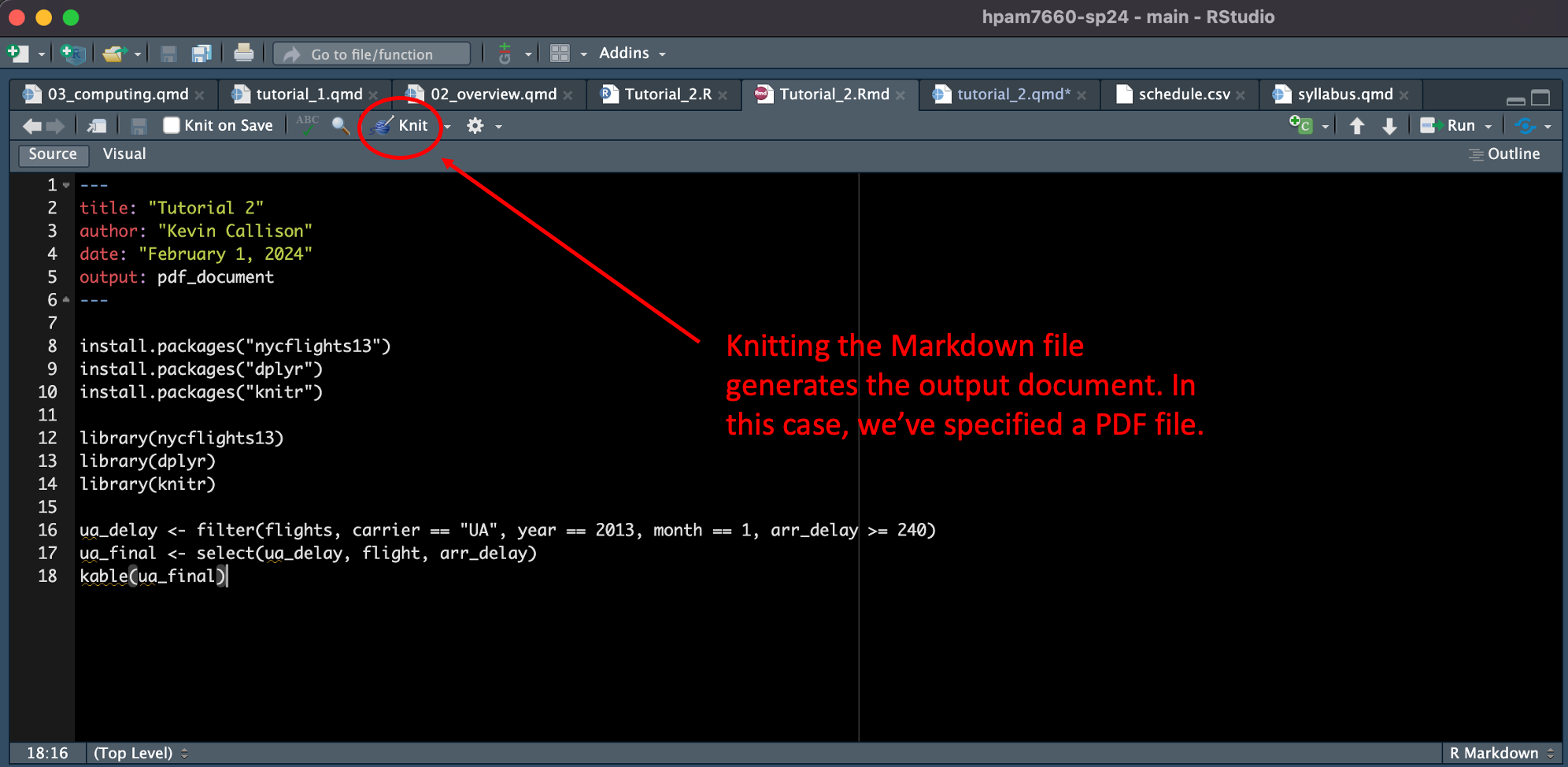
You should see a message in the “Render” tab telling you that the output has been created and now see a PDF file that includes the title, your name, the date, and the R code that you typed. Something like this:

This is good, but we can do better. For one thing, Markdown doesn’t recognize the R code we included as code - it just sees it as text. But we can tell Markdown that it is R code by adding a code chunk by adding chunk delimiters at the beginning and end of the code:
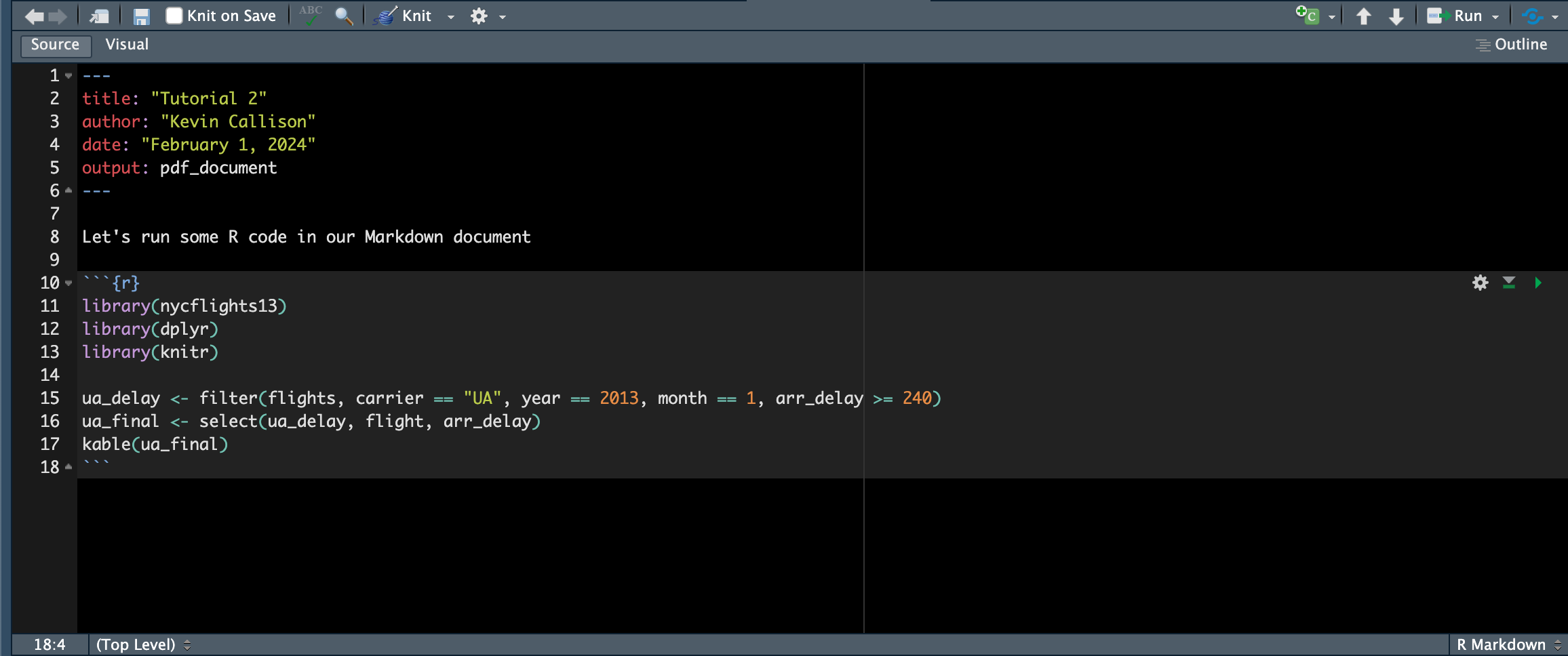
Now the PDF file contains our R code in nice grey blocks, along with any messages that R generates. More importantly, the PDF file also includes the output generated by the R commands.

It’s always good practice to annotate our code so that others know what we’re doing and why we’re doing it. Add some text to your Markdown files like I’ve done below:
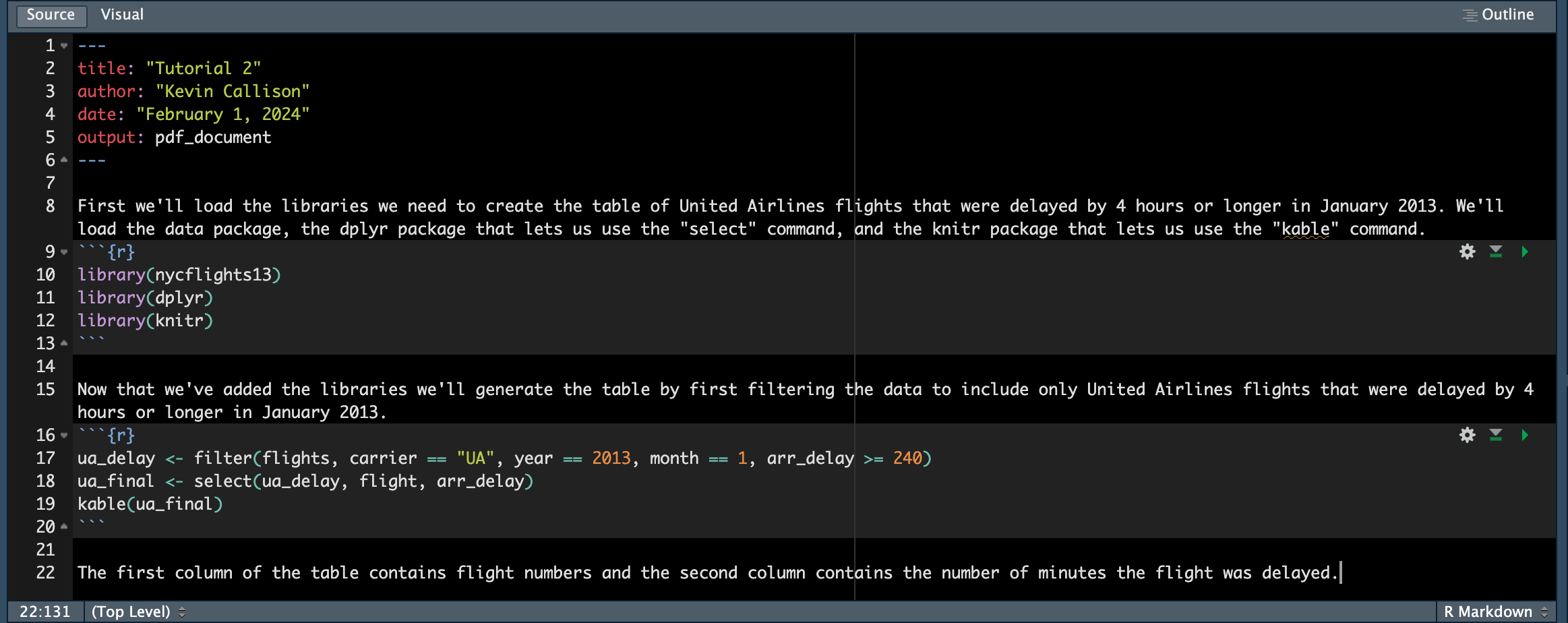
Your final result should look something like this:

We’re almost done, but we need to push our Markdown file to GitHub before we wrap up. If you click the “Git” tab in the upper right window, you should see the Markdown file listed. Now follow the same process we used to push our project files in Tutorial 1:
- Check the “Staged” box.
- Click the “Commit” button.
- Add a commit message. Maybe something like “Add Tutorial 2 Markdown File”.
- Click the “Commit” button.
- Close the Git Commit text box popup.
- Click the “Push” button.
- Close the Git Push popup.
Go to your GitHub page and make sure the file shows up in your hpam7660-tutorial2 repo. Invite me to the repo.
Congratulations! You made it all the way through!